7. Using BRIAN2 with pypet¶
7.1. pypet and BRIAN2¶
BRIAN2 as it comes is nice for small scripts and quick simulations, but it can be really hard to manage and maintain large scale projects based on very complicated networks with many parts and components. So I wrote a pypet extension that allows easier handling of more sophisticated BRIAN2 networks.
All of this can be found in pypet.brian2 sub-package.
The package contains a parameter.py file that includes specialized containers
for BRIAN2 data, like the Brian2Parameter,
the Brian2Result (both for BRIAN Quantities), and
the Brian2MonitorResult (extracts data from any kind of
BRIAN Monitor).
These can be used
in conjunction with the network management system in the network.py file within
the pypet.brian2 package.
In the following I want to explain how to use the network.py framework to run large
scale simulations. An example of such a large scale simulation can be found in
Large scale BRIAN2 simulation which is an implementation of the Litwin-Kumar and Doiron paper
from 2012.
7.2. The BRIAN2 network framework¶
The core idea behind my framework is that simulated spiking neural network are not in one giant piece but compartmentalize. Networks consist of NeuronGroups, Synapses, Monitors and so on and so forth. Thus, it would be neat if these parts can be easily replaced or augmented without rewriting a whole simulation. You want to add STDP to your network? Just plug-in an STDP component. You do not want to record anymore from the inhibitory neurons? Just throw away a recording component.
To abstract this idea, the whole simulation framework evolves around the
NetworkComponent class. This specifies an abstract API
that any component (which you as a user implement) should agree on to make them easy
to replace and communicate with each other.
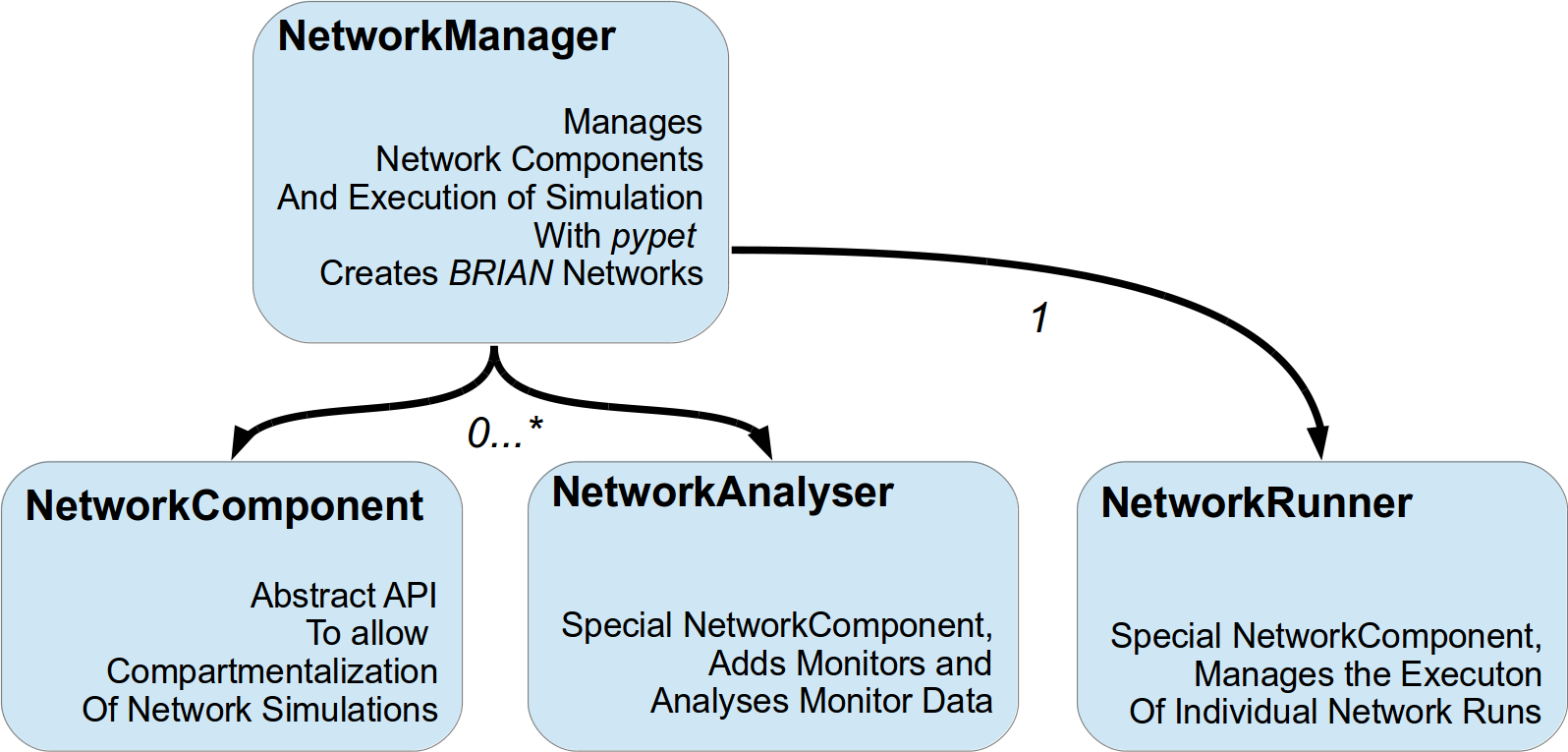
There are two specialisation of this NetworkComponent API:
The NetworkAnalyser and
the NetworkRunner. Implementations of the former deal with
the analysis of network output. This might range from simply adding and removing Monitors to
evaluating the monitor data and computing statistics about the network activity.
An instance of the latter is usually only created once and takes care about the running
of a network simulation.
All these three types of components are managed by the
NetworkManager that also creates BRIAN2 networks and
passes these to the runner.
Conceptually this is depicted in figure below.
7.3. Main Script¶
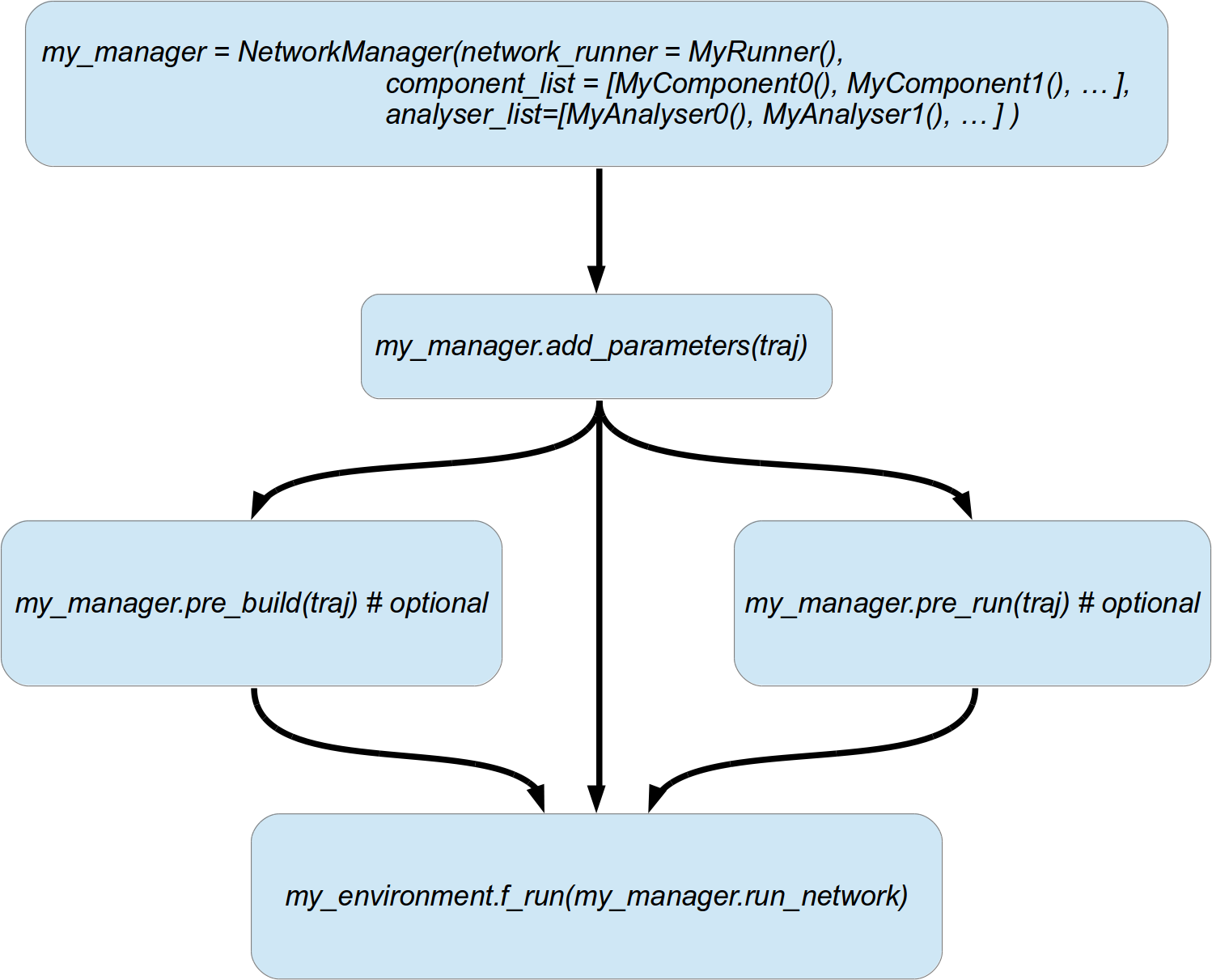
In your main script that you use to create an environment and start the parameter exploration, you also need to include these following steps.
- Create a
NetworkRunnerand your
NetworkComponentinstances andNetworkAnalyserinstances defining the layout and structure of your network and simulation.What components are and how to implement these will be discussed in the next section.
Create a
NetworkManager:Pass your
NetworkRunner(as first argument network_runner), all yourNetworkComponentinstances as a list (as second argumentcomponent_list) and allNetworkAnalyserinstances (as third argumentanalyser_list) to the constructor of the manager.Be aware that the order of components and analysers matter. The building of components, addition, removal, and analysis (for analyser) is executed in the order they are passed in the
component_listandanalyser_list, respectively. If a component B depends on A and C, make B appear after A and C in the list.For instance, you have an excitatory neuron group, an inhibitory one, and a connection between the two. Accordingly, your
NetworkComponentcreating the connection must be listed after the components responsible for creating the neuron groups.For now on let’s call the network manager instance
my_manager.Call
my_manager.add_parameters(traj):This automatically calls
add_parameters(traj)for all components, all analysers and the runner. So that they can add all their necessary parameters to the the trajectorytraj.(Optionally) call
my_manager.pre_build(traj):This will automatically trigger the
pre_buildfunction of your components, analysers and the network runner.This is useful if you have some components that do not change during parameter exploration, but which are costly to create and can be so in advance.
For example, you might have different neuron layers in your network and parts of the network do not change during the runtime of your simulation. For instance, your connections from an LGN neuron group to a V1 neuron group is fixed. Yet, the computation of the connection pattern is costly, so you can do this in
pre_buildto save some time instead of building these over and over again in every single run.(Optionally) call
my_manager.pre_run_network(traj)This will trigger a pre run of the network. First
my_manager.pre_buildis called (so you do not have to call it yourself if you intend a pre run). Then a novel BRIAN2 network instance is created from thebrian_list(see below). This network is simulated by your runner. The state after the pre run is preserved for all coming simulation runs during parameter exploration.This is useful if your parameter exploration does not involve modifications of the network per se. For instance, you explore different input stimuli which are tested on the very same network. Moreover, you have the very same initialisation run for every stimulus experiment. Instead of re-simulating the init run over and over again for every stimulus, you can perform it once as a pre run and use the network after the pre run for every stimulus input.
Pass the
run_network()to your environment’srun()to start parameter exploration. This will automatically initiate thebuild(traj)method for all your components, analysers and your runner in every single run. Subsequently, your network will be simulated with he help of your network runner.
These steps are also depicted in the figure below.
An example main script might look like the following:
from pypet.environment import Environment
from pypet.brian2.network import NetworkManager
from clusternet import CNMonitorAnalysis, CNNeuronGroup, CNNetworkRunner, CNConnections,\
CNFanoFactorComputer
env = Environment(trajectory='Clustered_Network',
add_time=False,
filename=filename,
continuable=False,
lazy_debug=False,
multiproc=True,
ncores=4,
use_pool=False, # We cannot use a pool, our network cannot be pickled
wrap_mode='QUEUE',
overwrite_file=True)
#Get the trajectory container
traj = env.trajectory
# We introduce a `meta` parameter that we can use to easily rescale our network
scale = 1.0 # To obtain the results from the paper scale this to 1.0
# Be aware that your machine will need a lot of memory then!
traj.f_add_parameter('simulation.scale', scale,
comment='Meta parameter that can scale default settings. '
'Rescales number of neurons and connections strenghts, but '
'not the clustersize.')
# We create a Manager and pass all our components to the Manager.
# Note the order, CNNeuronGroups are scheduled before CNConnections,
# and the Fano Factor computation depends on the CNMonitorAnalysis
clustered_network_manager = NetworkManager(network_runner=CNNetworkRunner(),
component_list=(CNNeuronGroup(), CNConnections()),
analyser_list=(CNMonitorAnalysis(),CNFanoFactorComputer()))
# Add original parameters (but scaled according to `scale`)
clustered_network_manager.add_parameters(traj)
# We need `tolist` here since our parameter is a python float and not a
# numpy float.
explore_list = np.arange(1.0, 3.5, 0.4).tolist()
# Explore different values of `R_ee`
traj.f_explore({'R_ee' : explore_list})
# Pre-build network components
clustered_network_manager.pre_build(traj)
# Run the network simulation
traj.f_store() # Let's store the parameters already before the run
env.run(clustered_network_manager.run_network)
# Finally disable logging and close all log-files
env.disable_logging()
7.3.1. Multiprocessing and Iterative Processing¶
The framework is especially designed to allow for multiprocessing and to
distribute parameter exploration of network simulations onto several cpus.
Even if parts of your network cannot be pickled, multiprocessing
can be easily achieved by setting use_pool=False for your
Environment.
Next, I’ll go a bit more into detail about components and finally you will learn which steps are involved in a network simulation.
7.4. Network Components¶
Network components are the basic building blocks of a pypet BRIAN experiment. There exist three types:
- Ordinary
NetworkComponentNetworkAnalyserfor data analysis and recordingNetworkRunnerfor simulation execution.
And these are written by YOU (eventually except for the network runner). The classes above are only abstract and define the API that can be implemented to make pypet’s BRIAN framework do its job.
By subclassing these, you define components that build and create BRIAN2 objects. For example, you could have your own ExcNeuronGroupComponent that creates a NeuronGroup of excitatory neurons. Your ExcNeuronSynapsesComponent creates BRIAN Synapses to make recurrent connections within the excitatory neuron group. These brian objects (NeuronGroup and Synapses) are then taken by the network manager to construct a BRIAN2 network.
Every component can implement these 5 methods:
This function should only add parameters necessary for your component to your trajectory
traj.
pre_build()and/orbuild()Both are very similar and should trigger the construction of objects relevant to BRIAN2 like NeuronGroups or Synapses. However, they differ in when they are executed. The former is initiated either by you directly (aka
my_manger.pre_build(traj)), or by a pre run (my_manager.pre_run_network(traj)). The latter is called during your single runs for parameter exploration, before the BRIAN2 network is simulated by your runner.The two methods provide the following arguments:
trajTrajectory container, you can gather all parameters you need from here.
brian_listA non-nested (!) list of objects relevant to BRIAN2.
Your component has to add BRIAN2 objects to this list if these objects should be added to the BRIAN2 network at network creation. Your manager will create a BRIAN2 network via
Network(*brian_list).
network_dictAdd any item to this dictionary that should be shared or accessed by all your components and which are not part of the trajectory container. It is recommended to also put all items from the
brian_listinto the dictionary for completeness.For convenience I suggest documenting the implementation of
buildandpre-buildand the other component methods in your subclass like the following. Use statements like Adds for items that are added to the list and dictionary and Expects for what is needed to be part of thenetwork_dictin order to build the current component.For instance:
brian_list:
Adds:
4 Connections, between all types of neurons (e->e, e->i, i->e, i->i)
network_dict:
Expects:
‘neurons_i’: Inhibitory neuron group
‘neurons_e’: Excitatory neuron group
Adds:
- ‘connections’ : List of 4 Connections,
between all types of neurons (e->e, e->i, i->e, i->i)
This method is called shortly before a subrun of your simulation (see below).
Maybe you did not want to add a BRIAN2 object directly to the
networkon its creation, but sometime later. Here you have the chance to do that.For instance, you have a SpikeMonitor that should not record the initial first subrun but the second one. Accordingly, you did not pass it to the
brian_listinpre_build()orbuild(). You can now add your monitor to thenetworkvia itsaddfunctionality, see the the BRIAN2 network class.The
add_to_network()relies on the following arguments
trajTrajectoy container
networkBRIAN2 network created by your manager. Elements can be added via add(…).
current_subrun
Brian2Parameterspecifying the very next subrun to be simulated. See next section for subruns.
subrun_listList of
Brian2Parameterobjects that are to be simulated after the current subrun.
network_dictDictionary of items shared by all components.
This method is analogous to
add_to_network(). It is called after a subrun (and after analysis, see below), and gives you the chance to remove items from a network.For instance, you might want to remove a particular BRIAN Monitor to skip recording of coming subruns.
Be aware that these functions can be implemented, but they do not have to be. If your custom component misses one of these, there is no error thrown. Instead, simply pass is executed (see the source code!).
7.4.1. NetworkAnalyser¶
The NetworkAnalyser is a subclass of an ordinary component.
It augments the component API by the function
analyse().
The very same parameters as for add_to_network() are
passed to the analyse function. As the name suggests, you can run some analysis here.
This might involve extracting data from monitors or computing statistics like Fano Factors, etc.
7.4.2. NetworkRunner¶
The NetworkRunner is another subclass of an ordinary component.
The given NetworkRunner does not define an API but
provides functionality to execute a network experiment.
There’s no need for creating your own subclass. Yet, I still suggest subclassing the
NetworkRunner, but just implement the
add_parameters() method. There you can add
Brian2Parameter instances to your trajectory
to define how long a network simulation lasts and in how many subruns it is divided.
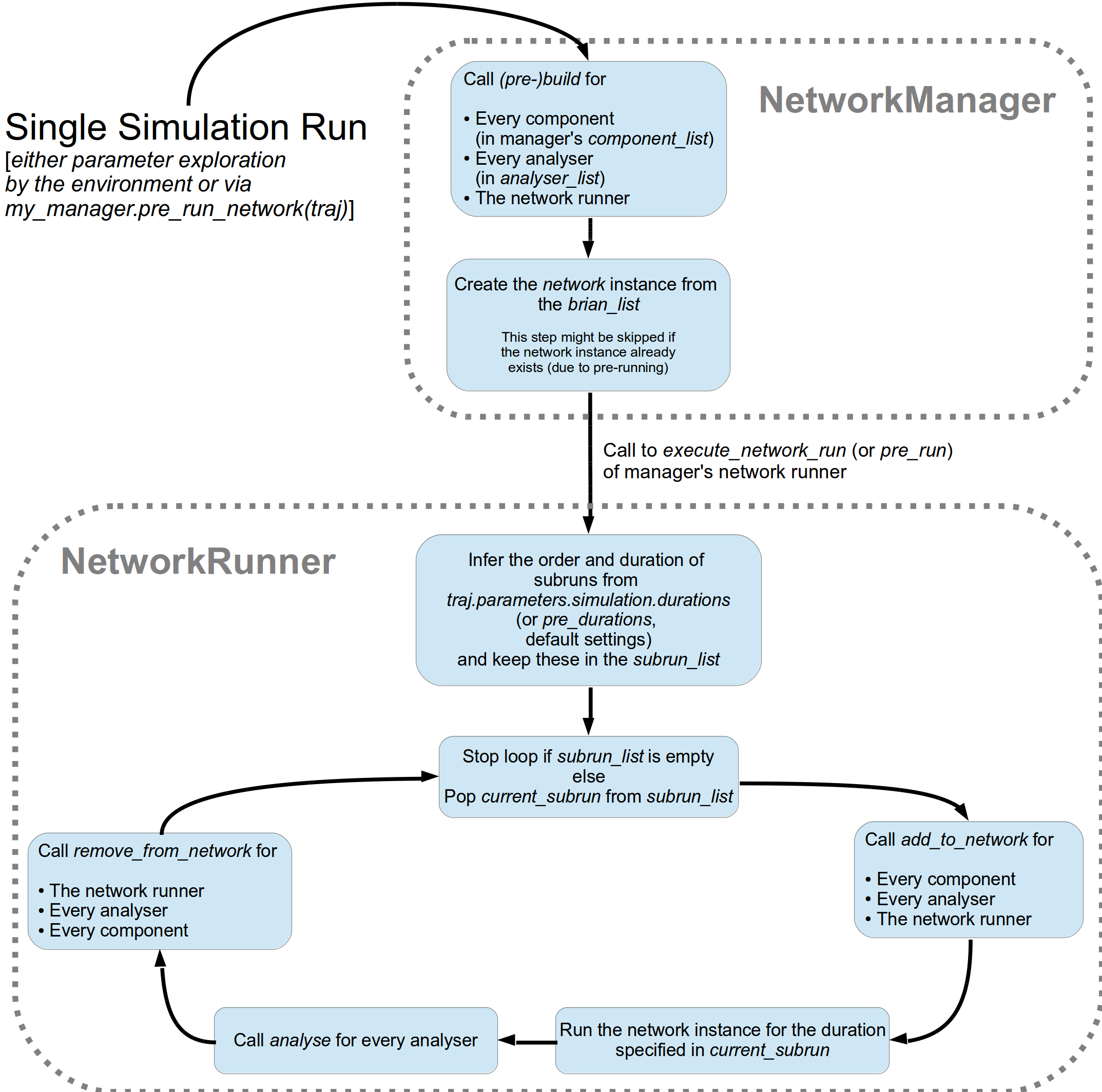
7.5. A Simulation Run and Subruns¶
A single run of a network simulation is further subdivided into so called subruns.
This holds for a pre run triggered by my_manager.pre_run_network(traj) as well
as an actual single run during parameter exploration.
The subdivision of a single run into further subruns is necessary to allow having
different phases of a simulation. For instance, you might want to run your network
for an initial phase (subrun) of 500 milliseconds. Then one of your analyser components checks for
pathological activity like too high firing rates. If this activity is detected, you
cancel all further subruns and skip the rest of the single run. You can do this by simply
removing all subruns from the subrun_list. You could also add further
Brian2Parameter instances to the list to make your
simulations last longer.
The subrun_list (as it is passed to add_to_network(),
remove_from_network(), or
analyse()) is populated by your network runner
at the beginning of every single run (or pre-run) in your parameter exploration.
The network runner searches for Brian2Parameter instances
in a specific group in your trajectory. By default this group is
traj.parameters.simulation.durations
(or traj.parameters.simulation.pre_durations for a pre-run),
but you can pick another group name when you create a NetworkRunner
instance. The order of the subruns is inferred from the v_annotations.order attribute of
the Brian2Parameter instances. The subruns are
executed in increasing order. The orders do not need to be consecutive, but a RuntimeError
is thrown in case two subruns have the same order. There is also an Error raised if there exists a
parameter where order cannot be found in it’s v_annotations property.
For instance, in traj.parameter.simulation.durations there are three
Brian2Parameter instances.
>>> init_run = traj.parameter.simulation.durations.f_add_parameter('init_run', 500 * ms)
>>> init_run.v_annotations.order=0
>>> third_run = traj.parameter.simulation.durations.f_add_parameter('third_run', 1.25 * second)
>>> third_run.v_annotations.order=42
>>> measurement_run = traj.parameter.simulation.durations.f_add_parameter('measurement_run', 15 * second)
>>> measurement_run.v_annotations.order=1
One is called init_run, has v_annotations.order=0 and lasts 500 milliseconds
(this is not cpu runtime but BRIAN simulation time).
Another one is called third_run lasts 1.25 seconds and has order 42.
The third one is named measurement_run lasts 5 seconds and has order 1.
Thus, a single run involves three subruns. They are executed in the order:
init_run involving running the network for
0.5 seconds, measurement_run for 5 seconds, and finally third_run for 1.25 seconds,
because 0 < 1 < 42.
The current_subrun Brian2Parameter
is taken from the subrun_list.
In every subrun the NetworkRunner will call
- for all ordinary components
- for all analysers
- for the network runner itself
run(duration)from the BRIAN2 network created by the manager.Where the
durationis simply the data handled by thecurrent_subrunwhich is aBrianParameter.
analyse()for all analysers
- for the network runner itself
- for all analysers
- for all ordinary components
The workflow of network simulation run is also depicted in the figure below.
I recommend taking a look at the source code in the pypet.brian2.network python file
for a better understanding how the pypet BRIAN framework can be used.
Especially, check the _execute_network_run()
method that performs the steps mentioned above.
Finally, despite the risk to repeat myself too much, there is an example on how to use pypet with BRIAN based on the paper by Litwin-Kumar and Doiron paper from 2012, see Large scale BRIAN2 simulation.
Cheers,
Robert