Naming Convention¶
To avoid confusion with natural naming scheme and the functionality provided by the trajectory, parameters, and so on, I followed the idea by PyTables to use prefixes: f_ for functions and v_ for python variables/attributes/properties.
For instance, given a result instance myresult, myresult.v_comment is the object’s comment attribute and myresult.f_set(mydata=42) is the function for adding data to the result container. Whereas myresult.mydata might refer to a data item named mydata added by the user.
More on Trajectories and Single Runs¶
Trajectory¶
For some example code on on topics discussed here see the 02 Natural Naming, Storage and Loading script.
The Trajectory is the container for all results and parameters (see More on Parameters and Results) of your numerical experiments. Throughout the documentation instantiated objects of the Trajectory class are usually labeled traj. Probably you as user want to follow this convention, because writing the not abbreviated expression trajectory all the time in your code can become a bit annoying after some time.
If you carry out an experiment and actively explore the parameter space, you will encounter a second class of top-level container called SingleRun. This one is derived or created by the original trajectory [1] and is used during the individual runs of your experiment. Single runs are not much different from trajectories except that they provide a little bit less functionality and only contain on particular parameter combination out of the full explored ranges. Since they are not much different from the original trajectory, within code they are also labeled with traj. We will come back to single runs later, but for now let’s focus on a trajectory.
The trajectory container instantiates a tree with groups and leaf nodes, whereas the trajectory object itself is the root node of the tree. There are two types of objects that can be leaves, parameters and results. Both follow particular APIs (see Parameter and Result as well as their abstract base classes BaseParameter, BaseResult). Every parameters contains a single value and optionally a range of values for exploration. In contrast, results can contain several heterogeneous data items (see More on Parameters and Results).
Moreover, a trajectory contains 4 major branches of its tree.
config
Parameters stored under config do not specify the outcome of your simulations but only the way how the simulations are carried out. For instance, this might encompass the number of cpu cores for multiprocessing. If you use and generate a trajectory with an environment (More about the Environment), the environment will add some config data to your trajectory.
Any leaf added under config is a Parameter object (or descendant of the corresponding base class BaseParameter).
As normal parameters, config parameters can only be specified before the actual single runs.
parameters
Parameters are the fundamental building blocks of your simulations. Changing a parameter usually effects the results you obtain in the end. The set of parameters should be complete and sufficient to characterize a simulation. Running a numerical simulation twice with the very same parameter settings should give also the very same results. Therefore, it is recommenced to also incorporate seeds for random number generators in your parameter set.
Any leaf added under parameters is a Parameter object (or descendant of the corresponding base class BaseParameter).
Parameters can only be introduced to the trajectory before the actual simulation runs.
derived_parameters
Derived parameters are specifications of your simulations that, as the name says, depend on your original parameters but are still used to carry out your simulation. They are somewhat too premature to be considered as final results. For example, assume a simulation of a neural network, a derived parameter could be the connection matrix specifying how the neurons are linked to each other. Of course, the matrix is completely determined by some parameters, one could think of some kernel parameters and a random seed, but still you actually need the connection matrix to build the final network.
Any leaf added under derived_parameters is a Parameter object (or descendant of the corresponding base class BaseParameter).
results
I guess results are rather self explanatory. Any leaf added under results is a Results object (or descendant of the corresponding base class BaseResult).
Note that all nodes provide the field ‘v_comment’, which can be filled manually or on construction via 'comment='. To allow others to understand your simulations it is very helpful to provide such a comment and explain what your parameter is good for. For parameters this comment will actually be shown in the parameter overview table (to reduce file size it is not shown in the result and derived parameter overview tables, see also Overview Tables). It can also be found as an HDF5 attribute of the corresponding nodes in the HDF5 file (this is true for all leaves).
| [1] | As a side remark, programming-wise the Trajectory class inherits from the SingleRun class. This yields a cleaner implementation than the other way round. |
Addition of Groups and Leaves (aka Results and Parameters)¶
Addition of leaves can be achieved via the functions:
Leaves can be added to any group, including the root group, i.e. the trajectory or the single run object themselves. Note that if you operate in the parameters subbranch of the tree, you can only add parameters (i.e. traj.parameters.f_add_parameter(...) but traj.parameters.f_add_result(...) does not work). For other subbranches this is analogous.
There are two ways to add these objects, either you already have an instantiation of the object, i.e. you add a given parameter:
>>> my_param = Parameter('subgroup1.subgroup2.myparam',42, comment='I am an example')
>>> traj.f_add_parameter(my_param)
Or you let the trajectory create the parameter, where the name is the first positional argument:
>>> traj.f_add_parameter('subgroup1.subgroup2.myparam', 42, comment='I am an example')
There exists a standard constructor that is called in case you let the trajectory create the parameter. The standard constructor can be changed via the v_standard_parameter property. Default is the Parameter constructor.
If you only want to add a different type of parameter once, but not change the standard constructor in general, you can add the constructor as the first positional argument followed by the name as the second argument:
>>> traj.f_add_parameter(PickleParameter, 'subgroup1.subgroup2.myparam', 42, comment='I am an example')
Derived parameters, config and results work analogously.
You can sort parameters/results into groups by colons in the names. For instance, traj.f_add_parameter('traffic.mobiles.ncars', data = 42) would create a parameter that is added to the subbranch parameters. This will also automatically create the subgroups traffic and inside there the group mobiles. If you add the parameter traj.f_add_parameter('traffic.mobiles.ncycles', data = 11) afterwards, you will find this parameter also in the group traj.parameters.traffic.ncycles.
Besides leaves you can also add empty groups to the trajectory (and to all subgroups, of course) via:
As before, if you create the group groupA.groupB.groupC and if group A and B were non-existent before, they will be created on the way.
Note that I distinguish between three different types of name, the full name which would be, for instance, parameters.groupA.groupB.myparam, the (short) name myparam and the location parameters.groupA.groupB. All these properties are accessible for each group and leaf via:
- v_full_name
- v_location
- v_name
Location and full name are relative to the root node, since a trajectory object (and single runs) is the root, it’s full_name is '' the empty string. Yet, the name property is not empty but contains the user chosen name of the trajectory.
Note that if you add a parameter/result/group with f_add_XXXXXX the full name will be extended by the full name of the group you added it to:
>>> traj.parameters.traffic.f_add_parameter('street.nzebras')
The full name of the new parameter is going to be parameters.traffic.street.nzebras. If you add anything directly to the root group, i.e. the trajectory object (or a single run), the group names parameters, config, derived_parameters will be automatically added (of course, depending on what you add, config, a parameter etc.).
If you add a result or derived parameters during a single run, the name will be changed to include the current name of the run.
For instance, if you add a result during a single run (let’s assume it’s the first run) like traj.f_add_result('mygroup.myresult', 42, comment='An important result'), the result will be renamed to results.runs.run_00000000.mygroup.myresult. Accordingly, all results (and derived parameters) of all runs are stored into different parts of the tree and are kept independent.
If this sorting does not really suit you, and you don’t want your results and derived parameters to be put in the sub-branches runs.run_XXXXXXXXX (with XXXXXXXX the index of the current run), you can make use of the wildcard character '$'. If you add this character to the name of your new result or derived parameter, pypet will automatically replace this wildcard character with the name of the current run.
For instance, if you add a result during a single run (let’s assume again the first one) via traj.f_add_result('mygroup.$.myresult', 42, comment='An important result') the result will be renamed to results.mygroup.run_00000000.myresult. Thus, the branching of your tree happens on a lower level than before. Even traj.f_add_result('mygroup.mygroup.$', myresult=42, comment='An important result') is allowed.
You can also use the wildcard character in the preprocessing stage. Let’s assume you add the following derived parameter BEFORE the actual single runs via traj.f_add_derived_parameter('mygroup.$.myparam', 42, comment='An important parameter'). If that happend DURING a single run '$' would be renamed to run_XXXXXXXX (with XXXXXXXX the index of the run). Yet, if you add the parameter BEFORE the single runs, '$' will be replaced by the placeholder name run_ALL. So your new derived parameter here is now called mygroup.run_All.myparam.
Why is this useful?
Well, this is in particular useful if you pre-compute derived parameters before the single runs which depend on parameters that might be explored in the near future.
For example you have parameter seed and n and which you use to draw a vector of random numbers. You keep this vector as a derived parameter. As long as you do not explore different seeds or values of n you can compute the random numbers before the single runs to save time. Now, if you use the '$' statement right from the beginning it would not make a difference if the following statement was executed during the pre-processing stage or during the single runs:
np.random.seed(traj.parameters.seed)
traj.f_add_derived_parameter('random_vector.$', np.random(traj.paramaters.n))
Accordingly, you have to write less code and post-processing and data analysis can become much easier.
Generic Addition¶
You do not have to stick to the given trajectory structure with its four subtrees: config, parameters, derived_parameters, results. If you just want to use a trajectory as a simple tree container and store groups and leaves wherever you like, you can use the generic functions f_add_group() and f_add_leaf(). Note however, that the four subtrees are reserved. Thus, if you add anything below one of the four, the corresponding speciality functions from above are called instead of the generic ones.
Note however, if you add any items during a single run, which are not located below a group called run_XXXXXXXX (where run_XXXXXXXXX is the name of your current run) these items are not automatically stored and you need to store them manually before the end of the run via f_store_items().
Accessing Data in the Trajectory¶
To access data that you have put into your trajectory you can use
- f_get() method. You might want to take a look at the function definition to check out the other arguments you can pass to f_get(). f_get not only works for the trajectory object, but for any group node in your tree.
- Use natural naming dot notation like traj.nzebras. This natural naming scheme supports some special features see below.
- Use the square brackets - as you do with dictionaries - like traj['nzebras'] which is similar to calling traj.nzebras.
Natural Naming¶
As said before trajectories instantiate trees and the tree can be browsed via natural naming.
For instance, if you add a parameter via traj.f_add_parameter('traffic.street.nzebras', data=4), you can access it via
>>> traj.parameters.street.nzebras
4
Here comes also the concept of fast access. Instead of the parameter object you directly access the data value 4. Whether or not you want fast access is determined by the value of v_fast_access (default is True):
>>> traj.v_fast_access = False
>>> traj.parameters.street.nzebras
<Parameter object>
Note that fast access works for parameter objects (i.e. for everything you store under parameters, derived_parameters, and config) that are non empty. If you say for instance traj.x and x is an empty parameter, you will get in return the parameter object. Fast access works in one particular case also for results, and that is, if the result contains exactly one item with the name of the result. For instance if you add the result traj.f_add_result('z',42), you can fast access it, since the first positional argument is mapped to the name ‘z’ (See also Results). If it is empty or contains more than one item you will always get in return the result object.
>>> traj.f_add_result('z', 42)
>>> traj.z
42
>>> traj.f_add_result('k', kay=42)
>>> traj.k
<Result object>
>>> traj.k.kay
42
>>> traj.f_add_result('two_data_values', 11, 12.0)
>>> traj.two_data_values
<Result object>
>>> traj.two_data_values[0]
11
Shortcuts¶
As a user you are encouraged to nicely group and structure your results as fine grain as possible. Yet, you might think that you will inevitably have to type a lot of names and colons to access your values and always state the full name of an item. This is, however, not true. There are two ways to work around that. First, you can request the group above the parameters, and then access the variables one by one:
>>> mobiles = traj.parameters.traffic.mobiles
>>> mobiles.ncars
42
>>> mobiles.ncycles
11
Or you can make use of shortcuts. If you leave out intermediate groups in your natural naming request, a breadth first search is applied to find the corresponding group/leaf.
>>> traj.mobiles
42
>>> traj.traffic.mobiles
42
>>> traj.parameters.ncycles
11
Search is established with very fast look up and usually needs much less then 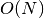 [most often
[most often 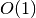 or
or 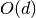 , where
, where 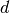 is the depth of the tree
and N the total number of nodes, i.e. groups + leaves].
is the depth of the tree
and N the total number of nodes, i.e. groups + leaves].
However, sometimes your shortcuts are not unique and you might find several solutions for your natural naming search in the tree. pypet will return the first item it finds via breadth first search within the tree. If there are several items with the same name but in different depths within the tree, the one with the lowest depth is returned. For performance reasons pypet actually stops the search if an item was found and there is no other item within the tree with the same name and same depth. If there happen to be two or more items with the same name and with the same depth in the tree, pypet will raise a NotUniqueNodeError since pypet cannot know which of the two items you want. [2]
The method that performs the natural naming search in the tree can be called directly, it is f_get().
>>> traj.parameters.f_get('mobiles.ncars')
<Parameter object ncars>
>>> traj.parameters.f_get('mobiles.ncars', fast_access=True)
42
If you don’t want to allow this shortcutting through the tree use f_get(target, shortcuts=False) or set the trajectory attribute v_shortcuts=False to forbid the shortcuts for natural naming and getitem access.
There also exist nice naming shortcuts for already present groups (these are always active and cannot be switched off):
- par is mapped to parameters, i.e. traj.parameters is the same group as traj.par
- dpar is mapped to derived_parameters
- res is mapped to results
- conf is mapped to config
- crun is mapped to the name of the current run (for example run_00000002)
- r_X and run_X are mapped to the corresponding run name, e.g. r_3 is mapped to run_00000003
For instance, traj.par.traffic.street.nzebras is equivalent to traj.parameters.traffic.street.nzebras.
Backwards search¶
Finally, there exists the possibility to perform bottom up search within the tree.
If you enable backwards search (set traj.v_backwards_search=True, default is False)
and use the square bracket notation or
f_get() and don’t pass a single name but a grouped
name separated via colons like
and using traj['groubA.groupB.paramC'] or
traj.f_get('groubA.groupB.paramC', backwards_search=True)
you can make pypet search the tree bottom up.
Thus, pypet won’t look for groupA first and than start looking for grougB from there and
finally search for paramC. But since it keeps internal indices and links to all it’s nodes
it will directly locate all entries within the tree named paramC and climb up the tree back
to the start node and
check if it passes by groupB and groupA on the way to the top.
Thus, the search complexity is
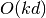 with
with 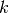 the number of occurrences of nodes named paramC and
the number of occurrences of nodes named paramC and
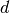 the depth of
your search tree. By the way, this backwards search always checks if your search term yields a
unique result irrespective of the depth of any of the nodes.
the depth of
your search tree. By the way, this backwards search always checks if your search term yields a
unique result irrespective of the depth of any of the nodes.
pypet will issue a performance warning if backwards search has to check too many terminal nodes. In this case you are advised to avoid shortcutting through the tree and state the full name of a parameter or result.
Note that backwards search is not triggered if the name can be directly found without shortcuts. For instance:
traj.f_add_parameters('groupA.groupB.paramC')
traj.v_backwards_search = True
traj['groupB.paramC'] # this will trigger backwards search
traj.parameters['groupA.groupB.paramC'] # this won't because
# 'parameters.groupA.groupB.paramC' is the real full name of the parameter
How is this backwards searching useful? Well, it will succeed in many more situations than simple breadth first forward traversal of the tree. For instance, let’s assume you have the following tree structure. traj.f_add_parameter('groupX.groupY.groupZ.paramA') adds a parameter paramA to your trajectory, similarly does traj.f_add_parameter('groupX.groupZ.paramB'). However, note the difference between the location of groupZ. These are in fact two different groups that have different depths in the trajectory tree! Now calling traj.groupZ.paramA will fail with an error, whereas traj['groupZ.paramA'] succeeds and will find paramA in your tree.
Why? traj.groupZ.paramA will initiate a breadth first forward tree traversal. To be precise, it will do so twice: At first pypet finds the group groupZ directly below groupX and, next, it tries to locate paramA from there. However, in groupX.groupZ it can only find paramB. Yet, if you enable backwards search and call traj['groupZ.paramA'], pypet directly looks for paramA and then moves up the tree back to the root note. It will find groupZ (the one below groupY) on the way and, therefore, knows that it has found the proper paramA.
Parameter Exploration¶
Exploration can be prepared with the function f_explore(). This function takes a dictionary with parameter names (not necessarily the full names, they are searched) as keys and iterables specifying how the parameter changes for each run as the values. Note that all iterables need to be of the same length. For example:
>>> traj.f_explore({'ncars':[42,44,45,46], 'ncycles' :[1,4,6,6]})
This would create a trajectory of length 4 and explore the four parameter space points
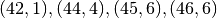 . If you want to explore the cartesian product of
parameter ranges, you can take a look
at the cartesian_product() function.
. If you want to explore the cartesian product of
parameter ranges, you can take a look
at the cartesian_product() function.
You can extend or expand an already explored trajectory to explore the parameter space further with the function f_expand().
Using Numpy Iterables¶
Note since parameters are very conservative regarding the data they accept (see Values supported by Parameters), you sometimes won’t be able to use Numpy arrays for exploration as iterables.
For instance, the following code snippet won’t work:
import numpy a np
from pypet.trajectory import Trajectory
traj = Trajectory()
traj.f_add_parameter('my_float_parameter', 42.4, comment='My value is a standard python float')
traj.f_explore( { 'my_float_parameter': np.arange(42.0, 44.876, 0.23) } )
This will result in a TypeError because your exploration iterable np.arange(42.0, 44.876, 0.23) contains numpy.float64 values whereas you parameter is supposed to use standard python floats.
Yet, you can use numpy’s tolist() function to overcome this problem:
traj.f_explore( { 'my_float_parameter': np.arange(42.0, 44.876, 0.23).tolist() } )
Or you could specify your parameter directly as a numpy float:
traj.f_add_parameter('my_float_parameter', np.float64(42.4),
comment='My value is a numpy 64 bit float')
Presetting of Parameters¶
I suggest that before you calculate any results or derived parameters, you should define all parameters used during your simulations. Usually you could do this by parsing a config file, or simply by executing some sort of a config file in python that simply adds the parameters to your trajectory (see also Tutorial).
If you have some complex simulations where you might use only parts of your parameters or you want to exclude a set of parameters and include some others, you can make use of the presetting of parameters (see pypet.trajectory.f_preset_parameter()). This allows you to add control flow on the setting or parameters. Let’s consider an example:
traj.f_add_parameter('traffic.mobiles.add_cars',True , comment='Whether to add some cars or '
'bicycles in the traffic simulation')
if traj.add_cars:
traj.f_add_parameter('traffic.mobiles.ncars', 42, comment='Number of cars in Rome')
else:
traj.f_add_parameter('traffic.mobiles.ncycles', 13, comment'Number of bikes, in case '
'there are no cars')
There you have some control flow. If the variable add_cars is True, you will add 42 cars otherwise 13 bikes. Yet, by your definition one line before add_cars will always be True. To switch between the use cases you can rely on presetting of parameters. If you have the following statement somewhere before in your main function, you can make the trajectory change the value of add_cars right after the parameter was added:
traj.f_preset_parameter('traffic.mobiles.add_cars', False)
So when it comes to the execution of the first line in example above, i.e. traj.f_add_parameter('traffic.mobiles.add_cars', True , comment='Whether to add some cars or bicycles in the traffic simulation')
The parameter will be added with the default value add_cars=True but immediately afterwards the pypet.parameter.Parameter.f_set() function will be called with the value False. Accordingly, if traj.add_cars: will evaluate to False and the bicycles will be added.
Note that in order to preset a parameter you need to state its full name (except the prefix parameters) and you cannot shortcut through the tree. Don’t worry about typos, before the running of your simulations it will be checked if all parameters marked for presetting were reached, if not a PresettingError will be thrown.
Storing¶
Storage of the trajectory container and all it’s content is not carried out by the trajectory itself but by a service. The service is known to the trajectory and can be changed via the v_storage_service property. The standard storage service (and the only one so far, you don’t bother write an SQL one? :-) is the HDF5StorageService. As a side remark, if you create a trajectory on your own (for loading) with the Trajectory class constructor and you pass it a filename, the trajectory will create an HDF5StorageService operating on that file for you.
You don’t have to interact with the service directly, storage can be initiated by several methods of the trajectory and it’s groups and subbranches (they format and hand over the request to the service). There is a general scheme to storage, which is whatever is stored to disk is the ground truth and therefore cannot/should not be changed. So basically as soon as you store parts of your trajectory to disk they will stay there! So far there is no real support for changing data that was stored to disk (you can delete or rewrite some of it, see below).
Why being so restrictive? Well, first of all, if you do simulations, they are like numerical scientific experiments, so you run them, collect your data and keep these results. There is usually no need to modify the first raw data after collecting it. You may analyse it and create novel results from the raw data, but you usually should have no incentive to modify your original raw data. Second of all, HDF5 is bad for modifying data which usually leads to fragmented HDF5 files and does not free memory on your hard drive. So there are already constraints by the file system used (but trust me this is minor compared to the awesome advantages of using HDF5, and as I said, why the heck do you wanna change your results, anyway?).
Just to state that again, if you save stuff to disk, it is set in stone! So if you modify data in RAM and store it again, the HDF5 storage service will simply ignore these modifications!
The most straightforward way to store everything is to say:
>>> traj.f_store()
and that’s it. In fact, if you use the trajectory in combination with the environment (see More about the Environment) you do not need to do this call by yourself at all, this is done by the environment.
If you store a trajectory to disk it’s tree structure is also found in the structure of the HDF5 file! In addition, there will be some overview tables summarizing what you stored into the HDF5 file. They can be found under the top-group overview, the different tables are listed in the Overview Tables section. By the way, you can switch the creation of these tables off passing the appropriate arguments to the Environment constructor to reduce the size of the final HDF5 file.
Storing data individually¶
More interesting is the approach to store individual items. Assume you computed a result that is extremely large. So you want to store it to disk, than free the result and forget about it for the rest of your simulation:
>>> large_result = traj.results.large_result
>>> traj.f_store_item(large_result)
>>> large_result.f_empty()
Note that in order to allow storage of single items, you need to have stored the trajectory at least once. If you operate during a single run, this has been done before, if not, simply call traj.f_store() once before. If you do not want to store anything but initialise the storage, you can pass the argument only_init=True, i.e. traj.f_store(only_init=True).
Moreover, if you call f_empty() on a large result, only the reference to the giant data block within the result is deleted. So in order to make the python garbage collector free the memory, you must ensure that you do not have any external reference of your own in your code to the giant data.
To avoid re-opening an closing of the HDF5 file over and over again there is also the possibility to store a list of items via f_store_items() or whole subtrees via f_store_child().
Loading¶
Sometimes you start your session not running an experiment, but loading an old trajectory. The first step in order to do that is to create a new empty trajectory - in case you have stored stuff into an HDF5 file, you can pass a filename to the Trajectory constructor - and call f_load() on it. You can also directly pass the filename to f_load() if you want to.
Give it a name or an index of the trajectory you want to select within the HDF5 file. For the index you can also count backwards, so -1 would yield the last or newest trajectory in an HDF5 file. If you don’t specify any of the two, the name of the current trajectory object is taken.
There are two load modes depending on the argument as_new
as_new=True
You load an old trajectory into your current one, and only load everything stored under parameters in order to rerun an old experiment. You could hand this loaded trajectory over to an Environment and carry out another the simulation again.
as_new=False
You want to load and old trajectory and analyse results you have obtained. The current name of your newly created trajectory will be changed to the name of the loaded one.
If you choose the latter load mode, you can specify how the individual subtrees config,*parameters*, derived_parameters, and results are loaded:
pypet.pypetconstants.LOAD_NOTHING: (0)
Nothing is loaded.
pypet.pypetconstants.LOAD_SKELETON: (1)
The skeleton is loaded including annotations (See Annotations). This means that only empty parameter and result objects will be created and you can manually load the data into them afterwards. Note that pypet.annotations.Annotations do not count as data and they will be loaded because they are assumed to be small.
pypet.pypetconstants.LOAD_DATA: (2)
The whole data is loaded. Note in case you have non-empty leaves already in RAM, these are left untouched.
pypet.pypetconstants.OVERWRITE_DATA: (3)
As before, but non-empty nodes are emptied and reloaded.
Compared to manual storage, you can also load single items manually via f_load_item(). If you load a large result with many entries you might consider loading only parts of it (see f_load_items()) Note in order to load a parameter, result or group, with f_load_item() it must exist in the current trajectory in RAM, if it does not you can always bring your skeleton of your trajectory tree up to date with f_update_skeleton(). This will load all items stored to disk and create empty instances. After a simulation is completed, you need to call this function to get the whole trajectory tree containing all new results and derived parameters.
And last but not least there is also f_load_child() in order to load whole subtrees.
Automatic Loading¶
The trajectory supports the nice feature to automatically loading data while you access it. Set traj.v_auto_load=True and you don’t have to care about loading at all during data analysis.
Enabling automatic loading will make pypet do two things. If you try to access group nodes or leaf nodes that are currently not in your trajectory on RAM but stored to disk, it will load these with data. Note that in order to automatically load data you cannot use shortcuts! Secondly, if your trajectory comes across an empty leaf node, it will load the data from disk (here shortcuts work again, since only data and not the skeleton has to be loaded).
For instance:
# Create the trajectory independent of the environment
traj = Trajectory(filename='./myfile.hdf5')
# We add a result
traj.f_add_result('mygropA.mygroupB.myresult', 42, comment='The answer')
# Now we store our trajectory
traj.f_store()
# We remove all results
traj.f_remove_child('results', recursive=True)
# We turn auto loading on
traj.v_auto_loading = True
# Now we can happily recall the result, since it is loaded while we access it.
# Stating `results` here is important. We removed the results node above, so
# we have to explicitly name it here to reload it, too. There are no shortcuts allowed
# for nodes that have to be loaded on the fly and that did not exist in memory before.
answer= traj.results.mygroupA,mygroupB.myresult
# And answer will be 42
# Ok next example, now we only remove the data. Since everything is loaded we can shortcut
# through the tree.
traj.f_get('myresult').f_empty()
# Btw we have to use `f_get` here to get the result itself and not the data `42` via fast
# access
# If we now access `myresult` again through the trajectory, it will be automatically loaded.
# Since the result itself is still in RAM but empty, we can shortcut through the tree:
answer = traj.myresult
# And again the answer will be 42
Logging and Git Commits during Data Analysis¶
Automated logging and git commits are often very handy features. Probably you do not want to miss these while you do your data analysis. To enable these in case you simply want to load an old trajectory for data analysis without doing any more single runs, you can again use an Environment.
First, load the trajectory with f_load(), and pass the loaded trajectory to a new environment. Accordingly the environment will trigger a git commit (in case you have specified a path to your repository root) and enable logging. You can additionally pass the argument do_single_runs=False to your environment if you only load your trajectory for data analysis. Accordingly, no config information like whether you want to use multiprocessing or resume a broken experiment is added to your trajectory. For example:
# Create the trajectory independent of the environment
traj = Trajectory(filename='./myfile.hdf5',
dynamically_imported_classes=[BrianParameter,
BrianMonitorResult,
BrianResult])
# Load the first trajectory in the file
traj.f_load(index=0, load_parameters=2,
load_derived_parameters=2, load_results=1,
load_other_data=1)
# Just pass the trajectory as the first argument to a new environment.
# You can pass the usual arguments for logging and git integration.
env = Environment(traj
log_folder='./logs/',
git_repository='../gitroot/',
do_single_runs=False)
# Here comes your data analysis...
Removal of items¶
If you only want to remove items from RAM (after storing them to disk), you can get rid of whole subbranches via f_remove_child().
But usually it is enough to simply free the data and keep empty results by using the f_empty() function of a result or parameter. This will leave the actual skeleton of the trajectory untouched.
Although I made it pretty clear that in general what is stored to disk is set in stone, there are a functions to delete items not only from RAM but also from disk: f_delete_item() and f_delete_items(). Note that you cannot delete explored parameters.
Merging and Backup¶
You can backup a trajectory with the function pypet.trajectory.Trajectory.f_backup().
If you have two trajectories that live in the same space you can merge them into one via pypet.trajectory.Trajectory.f_merge(). There are a variety of options how to merge them. You can even discard parameter space points that are equal in both trajectories. You can simply add more trials to a given trajectory if both contain a trial parameter. This is an integer parameter that simply runs from 0 to N1-1 and 0 to N2-1 with N1 trials in your current and N2 trials in the other trajectory, respectively. After merging the trial parameter in your merged trajectory runs from 0 to N1+N2-1.
Also checkout the example in 03 Merging of Trajectories.
Single Runs¶
As said before a SingleRun is like a smaller version of a trajectory. If you explore the parameter space, a single run is exactly one parameter space point that you visit on your trajectory during your numerical simulations. It is also the root node of your tree and offers slightly less functionality as the full trajectory.
How do you get single runs? They are the objects passed to your job functions. In 01 First Steps they are the traj parameter of the multiply function:
def multiply(traj):
z=traj.x*traj.y
traj.f_add_result('z', z, comment='Im the product of two values!')
As said before, they are not much different from trajectories, the best is you treat them as you would treat a trajectory object. Accordingly, the function argument is also named traj instead of singlerun.
A run is identified by it’s index and position in your trajectory, you can access this via v_idx. As a proper informatics nerd, if you have N runs, than your first run’s index is 0 and the last is indexed as N-1! Also each run has a name run_XXXXXXXX where XXXXXXXX is the index of the run with some leading zeros, like run_00000007.
Single run objects lack some functionality compared to trajectories:
You can no longer add config and parameters
- You can usually not access the full exploration range of parameters but only the current
value that corresponds to the index of the run.
Conceptually one should regard all single runs to be independent. As a consequence, you should NOT load data during a particular run that was computed by a previous one. You should NOT make a run manipulate data in the trajectory that was not added during the particular single run. This is very important! When it comes to multiprocessing, manipulating data put into the trajectory before the single runs is even more useless. Because the trajectory is either pickled or the whole memory space of the trajectory is forked by the OS, changing stuff within the trajectory will not be noticed by any other process or even the main script!
Interaction with Trajectories after an Experiment¶
Iterating over Loaded Data in a Trajectory¶
The trajectory offers a way to iteratively look into the data you have obtained from several runs. Assume you have computed the value z with z=traj.x*traj.x and added z to the trajectory/single run in each run via traj.f_add_result('z', z). Accordingly, you can find a couple of traj.results.runs.run_XXXXXXXX.z in your trajectory (where XXXXXXXX is the index of a particular run like 00000003). To access these one after the other it is quite tedious to write run_XXXXXXXX each time.
There is a way to tell the trajectory to only consider the subbranches that are associated with a single run and blind out everything else. You can use the function f_as_run() to make the trajectory only consider a particular run (it accepts run indices as well as names). Alternatively you can set the run idx via changing v_idx of your trajectory object. In addition to blinding out all branches that are not part of this run, all explored parameters within the trajectory are also set to the value associated with the corresponding index. Note that blinding out will also affect the functions f_iter_leaves() and f_iter_nodes().
In order to set everything back to normal call f_restore_default() or set v_idx to -1.
For example, consider your trajectory contains the parameters x and y and both have been
explored with 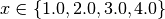 and
and 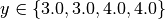 and
their product is stored as z. The following
code snippet will iterate over all four runs and print the result of each run:
and
their product is stored as z. The following
code snippet will iterate over all four runs and print the result of each run:
for run_name in traj.f_get_run_names():
traj.f_as_run(run_name)
x=traj.x
y=traj.y
z=traj.z
print '%s: x=%f, y=%f, z=%f' % (run_name,x,y,z)
# Don't forget to reset your trajectory to the default settings, to release its belief to
# be the last run:
traj.f_restore_default()
This will print the following statement:
run_00000000: x=1.000000, y=3.000000, z=3.000000
run_00000001: x=2.000000, y=3.000000, z=6.000000
run_00000002: x=3.000000, y=4.000000, z=12.000000
run_00000003: x=4.000000, y=4.000000, z=16.000000
To see this in action you might want to check out 03 Merging of Trajectories.
Looking for Subsets of Parameter Combinations (f_find_idx)¶
Let’s say you already explored the parameter space and gathered some results. The next step would be to post-process and analyse the results. Yet, you are not interested in all results at the moment but only for subsets where the parameters have certain values. You can find the corresponding run indices with the f_find_idx() function.
In order to filter for particular settings you need a lambda filter function and a list specifying the names of the parameters that you want to filter. You don’t know what lambda functions are? You might wanna read about it in Dive Into Python.
For instance, let’s assume we explored the parameters ‘x’ and ‘y’ and the cartesian product
of 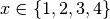 and
and 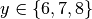 . We want to know the run indices for
x==2 or y==8. First we need to formulate a lambda filter function:
. We want to know the run indices for
x==2 or y==8. First we need to formulate a lambda filter function:
>>>my_filter_function = lambda x,y: x==2 or y==8
Next we can ask the trajectory to return an iterator over all run indices that fulfil the above named condition:
>>> idx_iterator = traj.f_find_idx(['parameters.x', 'parameters.y'],my_filter_function)
Note the list ['parameters.x', 'parameters.y'] to tell the trajectory which parameters are associated with the variables in the lambda function. Make sure they are in the same order as in your lambda function.
Now if we print the indexes found by the lambda filter, we get:
>>> print [idx for idx in idx_iterator]
[1, 5, 8, 9, 10, 11]
To see this in action check out 08 Using the f_find_idx Function.
Annotations¶
Annotations are a small extra feature. Every group node (including your trajectory, but not single runs) and every leaf has a property called v_annotations. These are other container objects (accessible via natural naming of course), where you can put whatever you want! So you can mark your items in a specific way beyond simple comments:
>>> ncars_obj = traj.f_get('ncars')
>>> ncars_obj.v_annotations.my_special_annotation = ['peter','paul','mary']
>>> print ncars_obj.v_annotations.my_special_annotation
['peter','paul','mary']
So here you added a list of strings as an annotation called my_special_annotation. These annotations map one to one to the attributes of your HDF5 nodes in your final hdf5 file. The high flexibility of annotating your items comes with the downside that storage and retrieval of annotations from the HDF5 file is very slow. Hence, only use short and small annotations. Consider annotations as a neat additional feature, but I don’t recommend using the annotations for large machine written stuff or storing large result like data (use the regular result objects to do that!).
For storage of annotations apply the same rule as for results and parameters, whatever is stored to disk is set in stone!
| [2] | In previous versions, pypet would stop immediately after the first encounter of a matching node. You had to force the lookup of unique matchings via v_check_uniqueness. This feature has been abolished since the behavior is inconsistent within different simulations. There is no ordering in nodes. So the children of a node are traversed arbitrarily since they are stored in dictionaries. Searching for one node could yield different results every time it was performed if two or more nodes happened to have the same name and were found within the same depth in the tree. Also in previous versions, you could choose depth first search instead of breadth first search. Yet, again since nodes are in arbitrary order, this search strategy is rather useless because the user cannot determine the traversal order of tree nodes. |